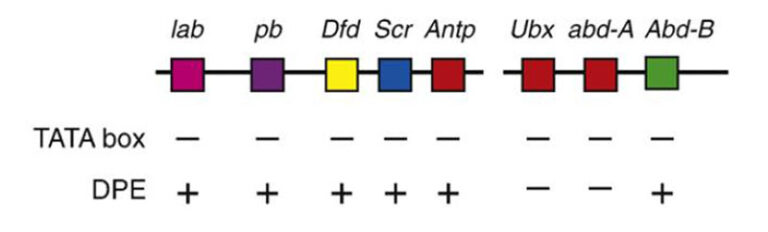
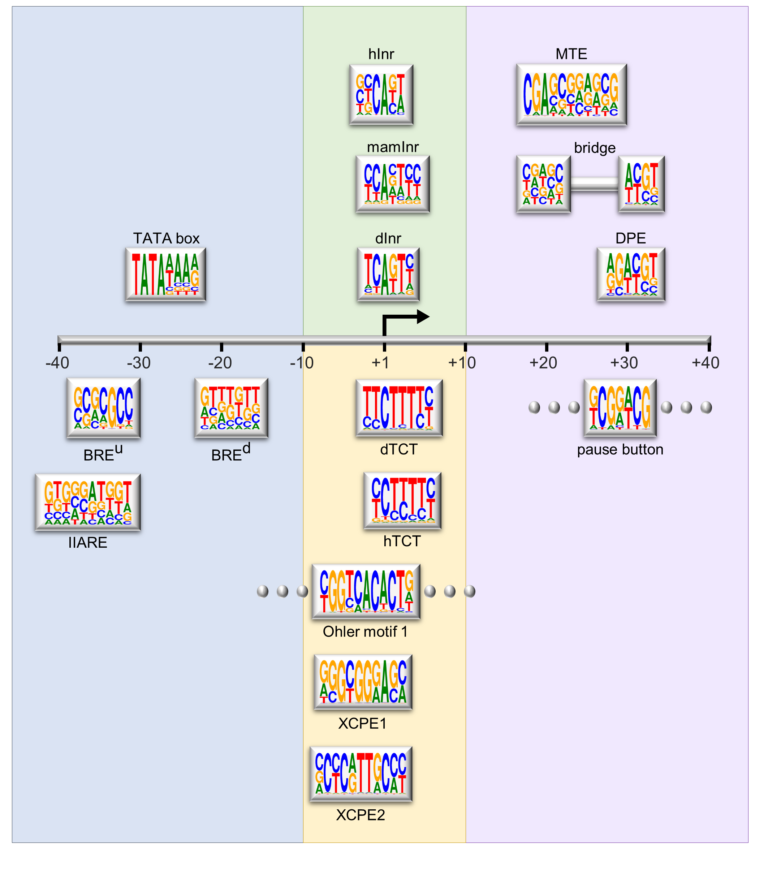
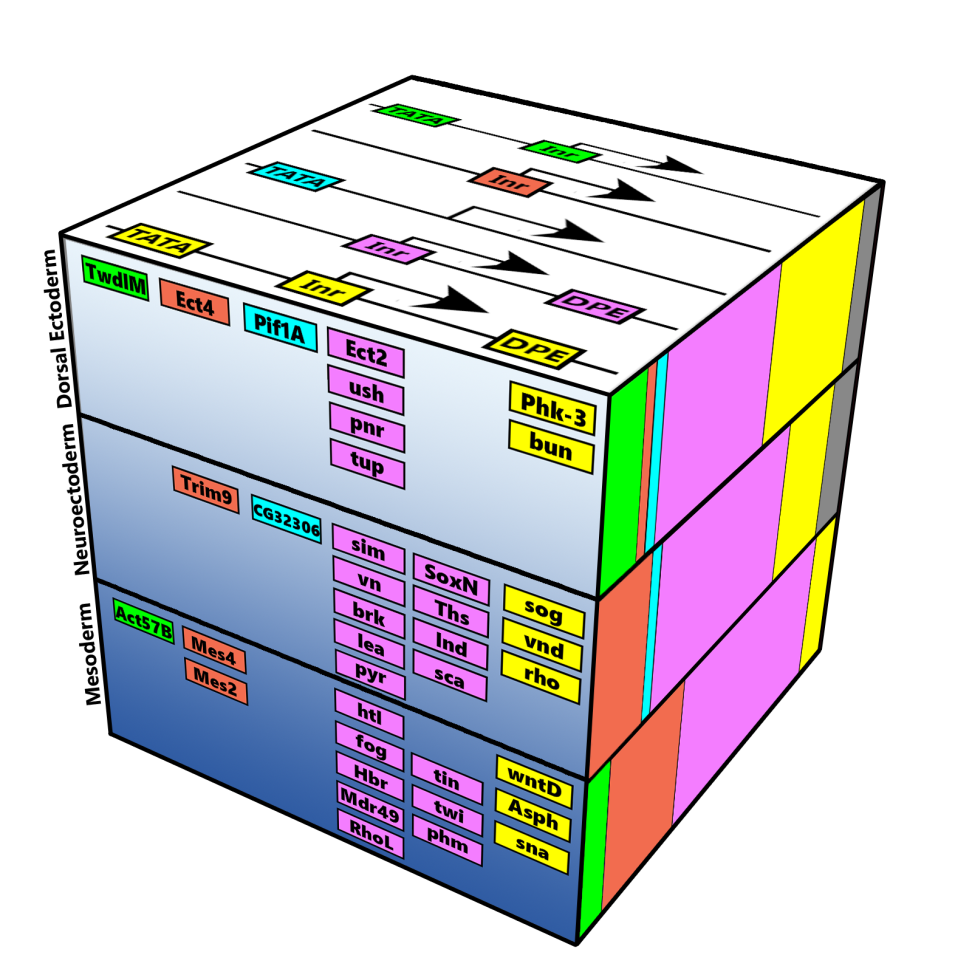
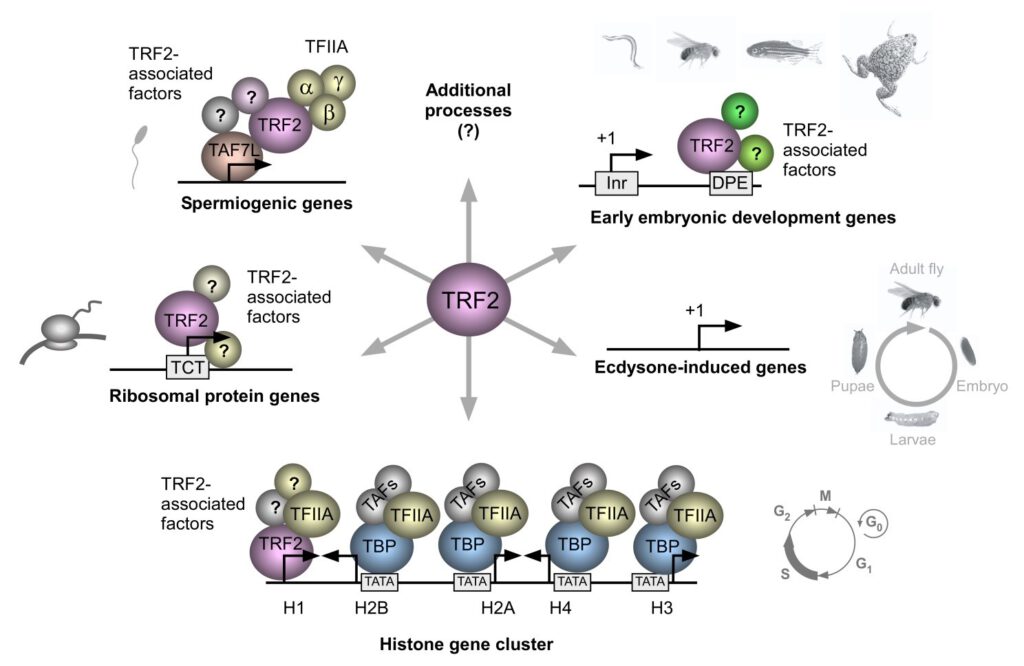
Schematic representation of the major core promoter elements.
Full information is available here
We study the molecular basis of the fascinating process by which our different body parts are formed during development.
The regulation of gene transcription is critical for the proper development and growth of an organism. The function of these genes is also crucial for us as adults; in the unfortunate event of malfunction, the end result might be cancer, such as leukemia.
Control elements that are embedded in the DNA sequences of genes are responsible for proper development. We study these DNA sequence elements and how they control different genes. Our analysis is important for understanding the regulation of development and complex systems.
We focus on the core promoter, which is generally defined as the DNA region that directs the accurate initiation of transcription by RNA polymerase II. In the past, the core promoter has often been presumed to be a generic entity that functions by a single universal mechanism. Recent findings reveal that there is widespread diversity in core promoter structure and function. We have embarked on the identification of biological functions of core promoter motifs and the identification of core promoter-specific activators.